
Dr Christophe RODRIGUEZ, Director
"Metagenomics, at the heart of screening and characterization of microbial pathogens."
The next-generation sequencing platform (NGS) was created in 2012 at the initiative of the Institute for Biomedical Research (IMRB), INSERM research center of the Faculty of Medicine of Créteil, and the Pôle of Biology and Pathology of the Henri Mondor Hospital Group (Assistance Publique-Hôpitaux de Paris), supported by INTER-DIM funding (Ile-de-France Region) and specific funding from the University Paris-Est Créteil, the Assistance Publique-Hôpitaux de Paris, and the National Agency for Research on AIDS and Viral Hepatitis (ANRS). The platform has focused on medical and microbiological research from the beginning and now occupies an important place in the development of NGS techniques for the diagnosis and characterization of microbial pathogens.
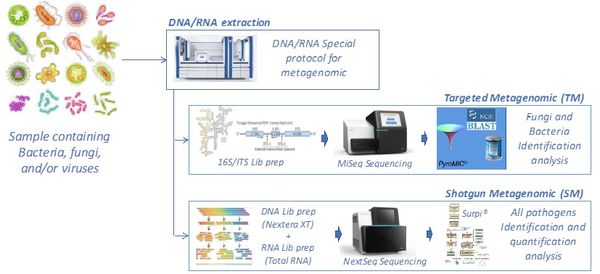
The constant emergence of new infections, favored by globalization, travel, and global warming, sometimes requires the diagnosis of infections based on nonspecific symptoms, in poorly informative contexts, without any preliminary knowledge of the microorganism. The development of specific diagnostic tests for a given infection is expensive, time-consuming to implement, and often available too late. The laboratory has specialized in the development and application of a "universal" diagnostic test based on a metagenomic approach. These metagenomic techniques aim to establish the repertoire of all the species present in a sample. The first application was targeted metagenomics (16S or ITS) for bacteria or fungi, but this approach did not allow the search for viruses, whether DNA or RNA.
Our laboratory has developed a complete process for "shotgun metagenomics" allowing:
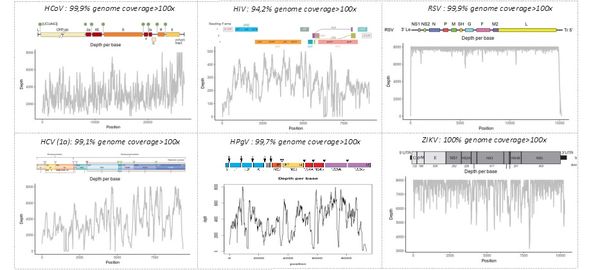
The "shotgun metagenomic" technique has been validated on mono- and polymicrobial samples containing bacterial, fungal, and/or viral pathogens. The figure below shows our ability to identify whole genome sequences of various viruses —including HIV, HCV, human pegivirus type 1 (HPGV), Zika virus (ZIKV), and respiratory syncytial virus (RSV)— in human samples or cell cultures (human coronavirus 229E, HCoV). The depth indicates the number of sequences generated for each region of the viral genome, allowing the identification of the pathogen by comparison with sequence libraries.
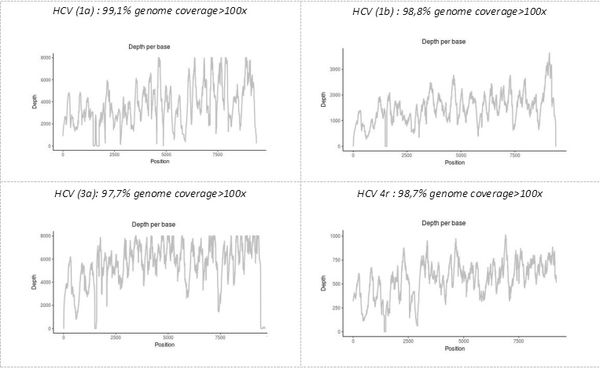
The precise identification of the pathogen(s), as well as demonstration that some of their genomic characteristics with functional significance, is another advantage of the "shotgun metagenomic" technique developed on the platform. Simultaneous sequencing of the complete genome of all pathogens present in a single sample makes it possible to obtain all the genetic data, including, possibly, those for minority variants. For example, the technique allows genotyping and subtyping or the detection of resistant viral variants in samples provided by the NRC (National Reference Center) of viral hepatitis B, C, and delta.
The platform also regularly tests new equipment and/or NGS kits as part of its technology monitoring mission, particularly for diagnosis and typing.
In the context of research, NGS techniques allow the characterization of modifications affecting the host in various infection situations, with or without anti-infectious treatment. The methods developed include RNA-Seq, micro-RNA sequencing, and transcriptomics. These tools provide valuable information for the identification of antiviral targets and the understanding of antiviral mechanisms of action for the group working on the development of new broad-spectrum antiviral approaches.
DNA, RNA extraction, all types of samplesQualitative and quantitative analysis of extracted nucleic acidsSequencing adapted to needs (MiSeq-NextSeq)