

PhD, MCU-PH, MD, PhD, PU-PH, PharmD
slim.fourati@aphp.fr stephane.chevaliez@aphp.fr
Clinical and translational research on the resistance of VHC to directly acting antivirals (DAAs)
The arrival of new antivirals against HCV on the market is creating the need for significant investment in the development of innovative techniques for two purposes: to search for amino acid substitutions associated with resistance (resistance-associated substitutions, RASs); and to determine the phenotypic consequences of these substitutions, especially on sensitivity to antivirals. The genotypic resistance tests we have established, and their interpretation, allow us to guide hepatologists, infectiologists, and all other clinicians who care for patients with chronic hepatitis C in their therapeutic decisions.
Our translational research focuses on the, sometimes complex, retreatment of patients who have failed treatment with DAAs and who were exposed to an inhibitor of the nonstructural protein NS5A. Such failures are usually associated with the selection of localized RASs in the NS5A protein sequence, most often conferring cross-resistance to all available NS5A protein inhibitors (daclatasvir, ledipasvir, ombitasvir, elbasvir, pibrentasvir and velpatasvir). These NS5A RASs do not alter the replicative capacity of most of the strains which carry them and can persist for several years, perhaps indefinitely, after stopping treatment, making retreatment problematic.
The general principles of retreatment are to use a combination based on sofosbuvir (nucleotide inhibitor of the protein NS5B, with a high barrier to resistance) and one or more other molecules, ideally without cross-resistance with that/those previously administered (EASL Recommendations for the Treatment of Hepatitis C 2016, J Hepatol 2017). Our center is particularly involved in the evaluation of new retreatment strategies for cases of AAD treatment multi-failure. For example, we recently characterized the resistance patterns observed in multi-failure patients receiving a triple combination with different classes of DAAs [sofosbuvir, daclatasvir, simeprevir (protease inhibitor NS3), and ribavirin]. The results showed the circulation of variants multi-resistant to different classes of antivirals (sometimes leading to a therapeutic impasse).
We are also studying the dynamics of DAA-resistant variants in pilot studies evaluating new therapeutic combinations in multi-failure patients, including:
Our second research topic is the optimization of treatment strategies, in terms of simplicity and duration, to improve patient adherence to treatment and possibly reduce costs. We have recently shown that those patients from a cohort of 221 cirrhotic patients (compensated cirrhosis without prior episode of decompensation) infected with HCV genotype 1 or 4, for whom no RAS was detectable before treatment (Sanger sequencing), could be effectively treated by 12 weeks of a ribavirin-free combination of sofosbuvir and an NS5A inhibitor. In contrast, this regimen was not effective in cirrhotic patients with NS5A RASs before treatment, suggesting that ribavirin is beneficial in these patients.
Phenotyping Platform for the Study of HCV Resistance to DAAs
The phenotyping platform for studying HCV resistance to AADs was set up in 2014 as part of the activities of the National Reference Center for Viral Hepatitis B, C, and Delta and the National Observatory of HCV Resistance to AADs, backed by the HEPATHER cohort and funded by the ANRS (ANRS CO22). This platform makes it possible to identify new patterns of DAA resistance to guide therapeutic strategies in cases of virological failure.
New amino-acid substitutions are introduced by site-directed mutagenesis in various subgenomic replicons (genotypes 1 to 5). The HCV replicon is based on a bicistronic chimeric construct organized as follows: a domain comprising the 5 'non-coding end of HCV, including an IRES (internal ribosome entry site), the neomycin-resistance gene (G418), a luciferase reporter gene, followed by a second domain comprising an EMCV (encephalomyocarditis virus) IRES which directs the translation of nonstructural HCV proteins, and ending with the 3' non-coding end of the viral genome. Subgenomic replicons lack the sequences that encode the structural proteins p7 and NS2, which are not necessary for their replication.
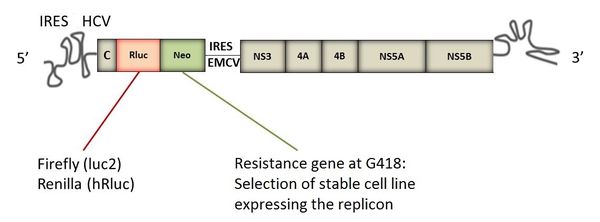
Structure of a subgenomic replicon of HCV
The candidate amino-acid substitutions are introduced by site-directed mutagenesis using one (or more) plasmid(s) and the phenotypic analysis is carried out using transient (replicons of subtype 1a, 1b and 2a) or so-called "stable" models"(replicates of genotype 3, 4, or 5) in the human hepatoma line, Huh-7.5.
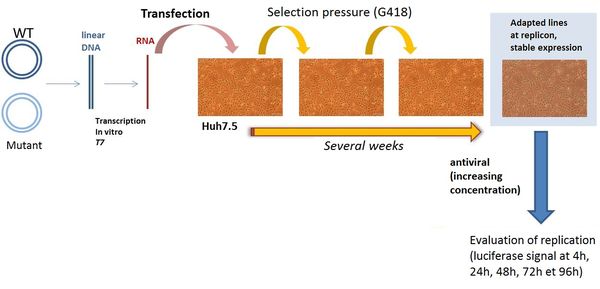
Measuring replication of a stable replicon model
After site-directed mutagenesis and linearization of the plasmid(s), it/they is/are transcribed in vitro by T7 RNA polymerase. The resulting subgenomic replicon RNA is transfected into Huh-7.5 cells. After stabilization (for genotypes 3, 4 or 5), phenotypic tests are performed in the presence of various concentrations of antivirals. The inhibition of viral replication is evaluated by measuring the luciferase activity in the presence of various concentrations of antivirals and at various post-treatment times. The resistance of the mutants is measured by assessing the 50% (EC 50) and the 90% effective concentrations (EC 90), defined as the antiviral concentrations that inhibit viral replication by 50 and 90%, respectively.
Analysis of the evolution dynamics of viral quasi-species under treatment and at the time of therapeutic failure by phylogenetic analysis of complete genomes using a shotgun metagenomic approach
Our group is studying the evolution of viral quasi-species by longitudinal analysis of HCV genome sequences (majority and minority) before, during, and after treatment (in cases of failure). This work, funded by the ANRS, aims to characterize the dynamics of viral quasi-species evolution during treatment and after its cessation (whole genome analysis by a "shotgun metagenomic" approach), and thus identify new substitutions outside the regions targeted by DAAs. The new substitutions will be introduced by site-directed mutagenesis into various replicons and tested against various antivirals to reveal the possible "compensation" or "resistance" they confer, as well as the replicative capacity of the corresponding replicons.
Phenotyping platform for the study of HBV sensitivity to antivirals
A platform for studying the phenotypic sensitivity of HBV to antivirals has recently been established in the laboratory. The objectives of this platform are to: (i) characterize substitutions associated with the resistance of HBV to various reverse transcriptase inhibitors and to molecules in therapeutic development and (ii) screen new molecules for inhibition of viral replication and/or other stages of the viral cycle.
The prototype is a plasmid encoding the genotype D, subtype 1 HBV genome under the control of the CMV promoter (Nassal et al, 1992).
Sofosbuvir-Daclatasvir-Simeprevir Plus Ribavirin in Direct-Acting Antiviral-Experienced Patients With Hepatitis C
Clin Infect Dis. 2017
Retreatment with sofosbuvir and simeprevir of patients with hepatitis C virus genotype 1 or 4 who previously failed a daclatasvir-containing regimen
Hepatology. 2016
Viral kinetics analysis and virological characterization of treatment failures in patients with chronic hepatitis C treated with sofosbuvir and an NS5A inhibitor
Alimentary Pharmacology Therapeutics 2017
Recent advances in understanding and diagnosing hepatitis B virus infection
F1000Res. 2016
Virologic Tools for HCV Drug Resistance Testing
Viruses. 2015